Drug repurposing is predicated on the fact that many diseases are caused by the dysregulation of similar signaling pathways, or that drugs may affect several biological targets at once, meaning that a single drug may be able to treat multiple diseases. Repurposing an existing drug is easier, cheaper and faster than developing a brand new drug, as there are comparatively few research and development costs, and the drug is already known to be effective and safe for use in patients.
Currently, the vast majority of additional indications for existing drugs have been found by chance (Liu et al., 2013). However, as technology improves, researchers have developed high throughput in silico methods to analyse the structures and biological activities of existing drugs to identify new indications (Ekins et al., 2011, Liu et al., 2013). A recent study by Gramatica et al. (2014) report an alternative approach, using computational linguistics and graph theory to identify previously unknown links between drugs and diseases.
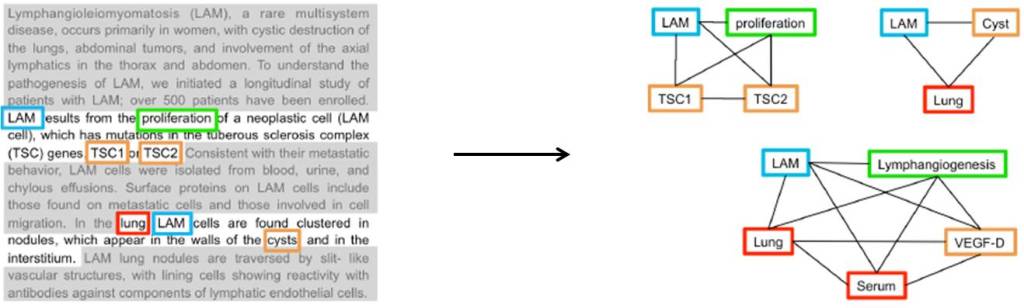
Gramatica et al. analysed three million publication abstracts relating to 300 rare diseases from the PubMed literature database. Analysing the language in the abstracts allowed the researchers to build visual networks linking different types of node, such as disease, protein, biological process, or drug. To show how these networks were built, the researchers used an example from a paper describing the pathogenesis of lymphangioleimyomatosis (LAM).
Each sentence was analysed independently, the nodes highlighted, and arranged into a network with each node linked pairwise, like so:
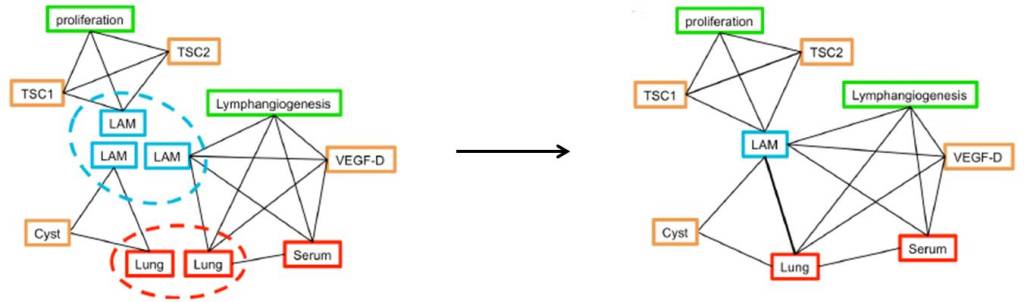
Each separate network was then rearranged to form a single, larger network representing the whole paper:
Where multiple links occur between nodes – such as LAM and Lung in this example – these nodes were considered to be more closely linked. Networks were then combined to cover multiple papers, and multiple diseases, building a large network of rare diseases, genes, biological processes, and drugs.
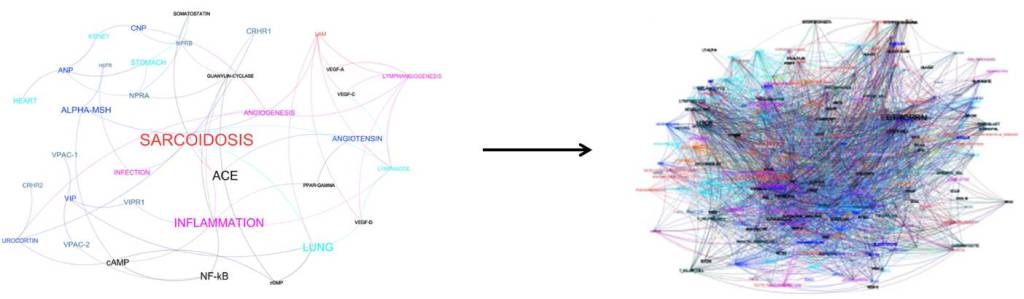
By arranging the results of a large number of studies – in this case three million – into a single network, links between drugs and diseases that were not previously recognized can be identified. Pathways with fewer steps linking a drug and a disease, and where multiple different pathways linked a drug and a disease, were considered to be particularly plausible candidates for drug repurposing.
To test whether this method led to the identification of new drug-disease pairs, the researchers used the network to identify new therapies for the rare lung disease Sarcoidosis, and new disease targets for the cancer drug Imatinib. They found that the peptides Aviptadil, ɑ-Melanocyte Stimulating Hormone (ɑ-MSH) and C-type Natriuretic Peptide (CNP) were possible candidates to treat Sarcoidosis. Indeed, a Phase II clinical trial in 20 patients reports that Aviptadil is an effective treatment for Sarcoidosis (Prasse et al., 2010), suggesting that ɑ-MSH and CNP may also be effective.
The network also predicted that Imatinib may be an effective treatment for spongiform encephalopathies, such as Creutzfeldt-Jakob disease. Imatinib inhibits the c-Abl tyrosine kinase, which has shown to be dysregulated and lead to neuronal cell death in multiple neurodegenerative disorders (Schlatterer et al., 2011) and Imatinib has been shown to clear mis-folded proteins from prion infected cells (Ertmer et al., 2004).
Together, these results suggest that combined computational linguistics and graph theory is able to identify previously unrecognized drug-disease pairs, which will expedite the repurposing of drugs for new indications.
Drug repurposing is a particularly valuable approach for identifying new treatments for rare and neglected diseases, where there is a high unmet medical need, and a number of rare disease organisations such as Findacure, Cures Within Reach and IRDiRC are actively funding and promoting this research. Indeed, there have been a number of successes in this area, for example Rapamycin has been repurposed as a treatment for Autoimmune Lymphoproliferative Syndrome and thalidomide is now an approved treatment for leprosy, multiple myeloma and bone marrow cancer (Teo et al., 2005).
The number of new therapies being approved for rare diseases is at an all-time high, but with only 133 new therapies approved since 2010, at this rate it will still take around 200 years to develop treatments for all rare diseases. Thus, although computational approaches to drug repurposing are in their infancy, the continued refinement of these approaches to expedite the discovery of drug-disease pairs will be of immense value to the field of rare diseases.
- Ekins S, Williams AJ, Krasowski MD, & Freundlich JS (2011). In silico repositioning of approved drugs for rare and neglected diseases. Drug discovery today, 16 (7-8), 298-310 PMID: 21376136
- Ertmer A, Gilch S, Yun SW, Flechsig E, Klebl B, Stein-Gerlach M, Klein MA, & Schätzl HM (2004). The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. The Journal of biological chemistry, 279 (40), 41918-27 PMID:15247213
- Gramatica R, Di Matteo T, Giorgetti S, Barbiani M, Bevec D, & Aste T (2014). Graph theory enables drug repurposing–how a mathematical model can drive the discovery of hidden mechanisms of action. PloS one, 9 (1) PMID: 24416311
- Liu Z, Fang H, Reagan K, Xu X, Mendrick DL, Slikker W Jr, & Tong W (2013). In silico drug repositioning: what we need to know.Drug discovery today, 18 (3-4), 110-5 PMID: 22935104
- Prasse A, Zissel G, Lützen N, Schupp J, Schmiedlin R, Gonzalez-Rey E, Rensing-Ehl A, Bacher G, Cavalli V, Bevec D, Delgado M, & Müller-Quernheim J (2010). Inhaled vasoactive intestinal peptide exerts immunoregulatory effects in sarcoidosis. American journal of respiratory and critical care medicine, 182 (4), 540-8 PMID: 20442436
- Schlatterer SD, Acker CM, & Davies P (2011). c-Abl in neurodegenerative disease. Journal of molecular neuroscience : MN, 45(3), 445-52 PMID: 21728062
- Teo SK, Stirling DI, & Zeldis JB (2005). Thalidomide as a novel therapeutic agent: new uses for an old product. Drug discovery today, 10 (2), 107-14 PMID: 15718159
www.bhdsyndrome.org – the primary online resource for anyone interested in BHD Syndrome.